Rectangular tree of R1a1a STR111 (as pdf):
Polar tree of R1a1a STR111 (as pdf):
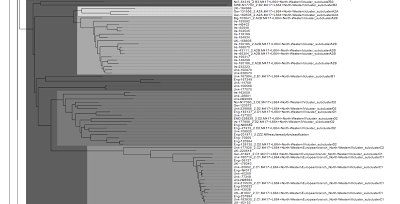
Due to the size of the tree I split the tree into pieces. Let's look into the details:
As mentioned the initial branching in the STR111 tree is pretty messy with no clear clustering but overlapping of various SNPs. At least N86494 from Belarus with M417- ended up in the pole position. Besides very small clusters from Ireland and England and a mix of Z93+ individuals (L657+ and L657-) from Central Asia and Middle East, the first clear cluster in this tree is "4. A2. Z283+ M458+ L260+ Central European branch, West Slavic subclade", most individuals are from Poland.
Next, the narrow "9. E1. Z93+ Z94+ Z2122+ "Ashkenazi-Levite" cluster" emerges. The closest isolate individual to this R1a1a Ashkenazi-Levite cluster is the Iraqi Kurdish individual H1483. I described this close genetic proximity in some earlier posts (here and here) when STR data were still very limited, but now we have STR111 data of H1483 confirming the predicted proximity.
Other close matches to the Ashkenazi-Levite cluster are 116213 from Palestine and two Scots (162927 and 221184).
On the next two parts, we have the Z280+ individuals forming a lot of clusters, one of them is the cluster "6. J1. Z280+ CTS1211+ (CTS3402+) Southern Baltic type". Within this large mega-cluster a few Z93+ isolated individuals and isolated clusters sneaked in, e.g. the "9. C7. Z93+ Z94+ L657- Z2122-, Arabic II" cluster, and the "9. C5. Z93+ Z94+ L657-(?), Bashkirs" cluster.
Next, we have no real clustering but a mess...
Next, the nice "Z280+ CT1211+ (CTS3402-) Carpathian cluster" comes up with a clear cut between P278+ and P278- individuals.
Next, the second group of M458+ individuals appears (L260- only) forming a nice cluster. This "Z283+ M458+ L1029+ L260-" is a Central European branch.
Next, we have a mix of individuals from both major branches (Z93 and Z283) followed by CTS3402+ individuals from Eastern Europe.
Next, we have a mix, no real clustering.
Next, we have Z284+ individuals forming a cluster:
Next, we have the L664+ individuals forming a large nice cluster. Based on the current R1a SNP tree, L664+ was a very early split. Most of these individuals are from the UK and Ireland.
Next, the Z92+ cluster emerges with people mostly from Eastern Europe.
Next, we have the Z284+ individuals forming two clusters, the Z288+/Z287+ cluster and a cluster of Z287- individuals.
Finally, the last cluster in this this tree: Z284+, L448+.
If you are getting confused by all these abbreviations, then this is okay. For clarification, I can recommend this schematic tree made by Michal.
http://eng.molgen.org/download/file.php?id=237&mode=view