Just a new update:
The individual that I described previously here turns out to be Z284-. So, he is still part of 'subclade 3' (see image below).
R1a1a (Z283+, only his paternal great-grandfather is Kurdish from Turkey):
The second individual with paternal Kurdish ancestry and the Y
chromosome haplogroup R1a1a is
definitely a very interesting case for Kurds as well as for the 'R1a1a
and Subclades Y chromosome project' itself, since he belongs to a
subclade (called subclade 3 in the project=R1a1a1g*) that was just
recently discovered and that split from the most common European
subclades in very ancient times. He is positive for Z283 and negative
for all tested underlying SNPs: Z283+ M417+ Z93- Z280- Pk5- P98- M64.2-
M56- M458- M434- M334- M157.1- L260- L176.1- L175- Z284-.
To get a better idea of R1a1a and its subclades (lineage tree image from the project):
1x E1b1b1c1a (Alevi)
1x G2a (Alevi)
2x J1 (Feyli, originally from Iran)
1x J1c3 (Sorani)
1x J2 (Kurd from Turkey)
1x J2a3a (J2a1a at 23andme; J2a4a at ISOGG 2009; he is M47+, M322+)(Yezidi)
1x T (Sorani)
1x R2a (Sorani)
1x R1b1a2* (Kurmanji from Zakho)
1x R1b1b2a (Zaza)
1x R1a1a (Sorani)
1x R1a1a (Z283+, subclade 3 only his paternal great-grandfather is Kurdish from Turkey)
1x I2a2a* (old I2b1*) (Sorani)
More data can be found here:
Kurdish Y-DNA Part I
Kurdish Y-DNA Part II
Kurdish Y-DNA Part IV
Kurdish Y-DNA Part V
Kurdish Y-DNA Part VI
Kurdish Y-DNA Part VII
Kurdish Y-DNA Part VIII
Total Pageviews
Wednesday, April 25, 2012
Monday, April 16, 2012
Cultural Distance Calculator
Today, I want to present work from Geert Hofstede, one of most influential persons in the research of national and organisational culture.
He came up with the idea to describe typical characteristics of cultures by only a few "cultural dimensions".
The Hofstede's cultural dimensions are described here, I will just quote very short descriptions of the dimensions (quotes from Hofstede's website).
1. Power distance index (PDI):
Fortunately, Hofstede provided values for the first 4 dimensions of 98 countries (in some cases he provided 5 dimensions) on his website.
Similar to genetic distance calculations based on genetic components, I calculated the distance of different societies based on the first 4 dimensions. You can download it here. The spreadsheet can be found here.
Update of this post can be found here.
He came up with the idea to describe typical characteristics of cultures by only a few "cultural dimensions".
The Hofstede's cultural dimensions are described here, I will just quote very short descriptions of the dimensions (quotes from Hofstede's website).
1. Power distance index (PDI):
- This dimension expresses the degree to which the less powerful members of a society accept and expect that power is distributed unequally.
- The high side of this dimension, called Individualism, can be defined as a preference for a loosely-knit social framework in which individuals are expected to take care of themselves and their immediate families only. Its opposite, Collectivism, represents a preference for a tightly-knit framework in society in which individuals can expect their relatives or members of a particular in-group to look after them in exchange for unquestioning loyalty.
- The masculinity side of this dimension represents a preference in society for achievement, heroism, assertiveness and material reward for success. Society at large is more competitive. Its opposite, femininity, stands for a preference for cooperation, modesty, caring for the weak and quality of life. Society at large is more consensus-oriented.
- The uncertainty avoidance dimension expresses the degree to which the members of a society feel uncomfortable with uncertainty and ambiguity. The fundamental issue here is how a society deals with the fact that the future can never be known: should we try to control the future or just let it happen? Countries exhibiting strong UAI maintain rigid codes of belief and behaviour and are intolerant of unorthodox behaviour and ideas. Weak UAI societies maintain a more relaxed attitude in which practice counts more than principles.
- The long-term orientation dimension can be interpreted as dealing with society’s search for virtue. Societies with a short-term orientation generally have a strong concern with establishing the absolute Truth. They are normative in their thinking.
- Indulgence stands for a society that allows relatively free gratification of basic and natural human drives related to enjoying life and having fun. Restraint stands for a society that suppresses gratification of needs and regulates it by means of strict social norms.
Fortunately, Hofstede provided values for the first 4 dimensions of 98 countries (in some cases he provided 5 dimensions) on his website.
Similar to genetic distance calculations based on genetic components, I calculated the distance of different societies based on the first 4 dimensions. You can download it here. The spreadsheet can be found here.
Update of this post can be found here.
Prominent nose!
Today, I want to share some thoughts, actually a chain of thoughts about how very few hormonal changes may effect various aspects of the phenotype.
One of the most visible phenotypical differences between East- and West Eurasians are the eyes (epicanthic fold of many East Eurasians) and the nose ('Bigger' nose of West Eurasians compared to East Eurasians). The latter one is the topic of my post.
So what makes the nose grow big? Besides embryonic development, most guys with a big nose know the answer: puberty.
Puberty is characterized by a hormonal change maturing a child body to an adult body. For men, this means change of the voice, beard growth, body hair, and nose growth. All these mayor phenotypical changes are mainly caused by one molecule, Testosterone (and its derivative Dihydrotestosterone or short DHT) .
Testosterone and DHT are hormones produced in the testes (in the Leydig cells), so both are messengers, transmitted as a little molecule from one part of the body (testis) to another part of the body (e.g. nose, hair follicle, etc.) via the blood vessels (to be precise, the skin can produce DHT by converting Testosterone to DHT). To receive and 'read' this transmitted molecular information, these tissues express a specific receptor called Androgen receptor. The timing, time duration and the strength of the information (testosterone concentration in blood) as well as the sensitivity/number of the Androgen receptors contributes to final outcome of the phenotype.
For example, baldness is associated with a mutation in the Androgen receptor causing hypersensitivity, which makes body hair darker and longer, and hair on the scalp progressively thinner after adolescence. The Androgen receptor is located on the X-chromosome, so this baldness-causing mutation is inherited from the mother (recently, mutations on other chromosomes has been associated with baldness, too). More severe mutations in the gene of the Androgen receptor can cause a loss of function (Complete androgen insensitivity syndrome or CAIS). Male individuals with such a non-functional Androgen receptor have a female appearance and different from both men and women, they mostly lack secondary terminal hair (e.g. underarm hair).
To get an better idea where the Androgen receptor is present and most likely crucial for phenotypical outcome, I can recommend this website: http://biogps.org/#goto=genereport&id=367
Higher expression (and most likely more presence) of the Androgen receptor can be found in various tissues, most of them are important for sexual differentiation of men and women: skin, Deltoid muscle, heart atrium, Skeletal muscles, in the epithelium of the Fallopian tube, Adipose tissue, breast (Breast ductal cells, breast lobular cells, mammary glands, nipple cross section), Penis, Prostate glands, Vulva, Vagina, Urethra, Cervix, Ovary, etc.
So, do men with more body hair have a more prominent nose? Viewed from a global perspective, I would say yes but I don't have any data for that.
Does the sexual dimorphism differ between the ethnic groups? Viewed from a global perspective, I would say yes but I don't have any data for that, either. I believe that sexual dimorphism is more pronounced in West Eurasians than in East Eurasians.
I believe that men with a more prominent nose are perceived as more dominant, similar to men with more facial hair. You might heard of the term 'Aristocratic nose', which is basically describing a bigger nose. And maybe big-nosed men are more dominant because Testosterone and the Androgen receptor are also present in the brain tissue where both together play an important role in behavioral outcome (e.g. aggression).
So, maybe there is a correlation between hormonal regulation, behavior, and 'nose' phenotype.
So where did the 'prominent nose people' first emerge? Evolutionary speaking, it is believed that narrow noses are an adaption to colder climate, and bigger noses are an adaption to warmer climate, so the first people with a more prominent nose must have been living in a warmer climate. Interestingly, ethnic groups speaking Indo-European and Semitic languages tend to have more people with prominent noses.
One of the most visible phenotypical differences between East- and West Eurasians are the eyes (epicanthic fold of many East Eurasians) and the nose ('Bigger' nose of West Eurasians compared to East Eurasians). The latter one is the topic of my post.
So what makes the nose grow big? Besides embryonic development, most guys with a big nose know the answer: puberty.
Puberty is characterized by a hormonal change maturing a child body to an adult body. For men, this means change of the voice, beard growth, body hair, and nose growth. All these mayor phenotypical changes are mainly caused by one molecule, Testosterone (and its derivative Dihydrotestosterone or short DHT) .
Testosterone and DHT are hormones produced in the testes (in the Leydig cells), so both are messengers, transmitted as a little molecule from one part of the body (testis) to another part of the body (e.g. nose, hair follicle, etc.) via the blood vessels (to be precise, the skin can produce DHT by converting Testosterone to DHT). To receive and 'read' this transmitted molecular information, these tissues express a specific receptor called Androgen receptor. The timing, time duration and the strength of the information (testosterone concentration in blood) as well as the sensitivity/number of the Androgen receptors contributes to final outcome of the phenotype.
For example, baldness is associated with a mutation in the Androgen receptor causing hypersensitivity, which makes body hair darker and longer, and hair on the scalp progressively thinner after adolescence. The Androgen receptor is located on the X-chromosome, so this baldness-causing mutation is inherited from the mother (recently, mutations on other chromosomes has been associated with baldness, too). More severe mutations in the gene of the Androgen receptor can cause a loss of function (Complete androgen insensitivity syndrome or CAIS). Male individuals with such a non-functional Androgen receptor have a female appearance and different from both men and women, they mostly lack secondary terminal hair (e.g. underarm hair).
To get an better idea where the Androgen receptor is present and most likely crucial for phenotypical outcome, I can recommend this website: http://biogps.org/#goto=genereport&id=367
Higher expression (and most likely more presence) of the Androgen receptor can be found in various tissues, most of them are important for sexual differentiation of men and women: skin, Deltoid muscle, heart atrium, Skeletal muscles, in the epithelium of the Fallopian tube, Adipose tissue, breast (Breast ductal cells, breast lobular cells, mammary glands, nipple cross section), Penis, Prostate glands, Vulva, Vagina, Urethra, Cervix, Ovary, etc.
So, do men with more body hair have a more prominent nose? Viewed from a global perspective, I would say yes but I don't have any data for that.
Does the sexual dimorphism differ between the ethnic groups? Viewed from a global perspective, I would say yes but I don't have any data for that, either. I believe that sexual dimorphism is more pronounced in West Eurasians than in East Eurasians.
I believe that men with a more prominent nose are perceived as more dominant, similar to men with more facial hair. You might heard of the term 'Aristocratic nose', which is basically describing a bigger nose. And maybe big-nosed men are more dominant because Testosterone and the Androgen receptor are also present in the brain tissue where both together play an important role in behavioral outcome (e.g. aggression).
So, maybe there is a correlation between hormonal regulation, behavior, and 'nose' phenotype.
So where did the 'prominent nose people' first emerge? Evolutionary speaking, it is believed that narrow noses are an adaption to colder climate, and bigger noses are an adaption to warmer climate, so the first people with a more prominent nose must have been living in a warmer climate. Interestingly, ethnic groups speaking Indo-European and Semitic languages tend to have more people with prominent noses.
Wednesday, April 11, 2012
Kurdish verbs
I just want to share one observation I made about Kurdish verbs. Quiet a few verbs in Sorani-Kurdish are combinations of a 'main verb' and a second word.
To give you an idea what I mean, here a list of Sorani-Verbs; in some cases the verb was obviously shortened. I am writing the Sorani-Kurdish words in Latin script (Sorani is usually written in Arabic script) , so the proper spelling might be a little bit off.
Bida! Give!
Sar Bida! Visit!
Pishani (Bi)da! Show!
Para Bida! Pay!
Iarmati Bida! Help!
Haul Bida! Try!
Zuab Bida! Respond!
Baz Bida! Jump!
Example #2:
Bika! Do!
Tatsruba Bika! Test!
Supas Bika! Thank!
Safar Bika! Travel!
Pirsiar Bika! Ask!
Peida Bika! Find!
Malla Bika! Swim!
Ish/Kar Bika! Work!
Iari Bika! Play!
Daua Bika! Order!
Dast Pe Bika! Start!
Cauare Bika! Expect!
Bir Bika! Think!
Bauir Bika! Believe!
Caki (Bi)ka! Fix!
Chrap (B)ika! Brake!
Tekall (B)ika! Mix!
Tam (B)ika! Taste!
Seir (B)ika! Look!
Ra (Bi)ka! Run!
Drust (B)ika! Make!
Fer (B)ika! Teach!
Example #3:
(U)asta! Stand!
Ra uasta! Stand (still)!
Halsta! Stand up!
Buasta! Wait!
Example #4:
Bigra! Grab! Hold!
Uar (b)igra! Receive!
Hal (b)igra! Pick up!
Ra (b)igra! Stop!
Gue bigra! Listen!
Example #5:
Biga! Arrive!
Te biga! Understand!
Example #6:
Biba! Be! Become!
Fer Ba! Learn!
Some interesting aspects of this exercise:
The stem of the word 'to run' is 'ra' in Sorani Kurdish.
Ra (Bi)ka! Run! (literally "Do run!")
Ra (b)igra! Stop! (literally "Hold run!")
Ra uasta! Stand (still)! (literally "Stand run!")
My personal favorite is:
Dast Pe Bika! Start! (literally "Do hand foot!") :-)
To give you an idea what I mean, here a list of Sorani-Verbs; in some cases the verb was obviously shortened. I am writing the Sorani-Kurdish words in Latin script (Sorani is usually written in Arabic script) , so the proper spelling might be a little bit off.
Bida! Give!
Sar Bida! Visit!
Pishani (Bi)da! Show!
Para Bida! Pay!
Iarmati Bida! Help!
Haul Bida! Try!
Zuab Bida! Respond!
Baz Bida! Jump!
Example #2:
Bika! Do!
Tatsruba Bika! Test!
Supas Bika! Thank!
Safar Bika! Travel!
Pirsiar Bika! Ask!
Peida Bika! Find!
Malla Bika! Swim!
Ish/Kar Bika! Work!
Iari Bika! Play!
Daua Bika! Order!
Dast Pe Bika! Start!
Cauare Bika! Expect!
Bir Bika! Think!
Bauir Bika! Believe!
Caki (Bi)ka! Fix!
Chrap (B)ika! Brake!
Tekall (B)ika! Mix!
Tam (B)ika! Taste!
Seir (B)ika! Look!
Ra (Bi)ka! Run!
Drust (B)ika! Make!
Fer (B)ika! Teach!
Example #3:
(U)asta! Stand!
Ra uasta! Stand (still)!
Halsta! Stand up!
Buasta! Wait!
Example #4:
Bigra! Grab! Hold!
Uar (b)igra! Receive!
Hal (b)igra! Pick up!
Ra (b)igra! Stop!
Gue bigra! Listen!
Example #5:
Biga! Arrive!
Te biga! Understand!
Example #6:
Biba! Be! Become!
Fer Ba! Learn!
Some interesting aspects of this exercise:
The stem of the word 'to run' is 'ra' in Sorani Kurdish.
Ra (Bi)ka! Run! (literally "Do run!")
Ra (b)igra! Stop! (literally "Hold run!")
Ra uasta! Stand (still)! (literally "Stand run!")
My personal favorite is:
Dast Pe Bika! Start! (literally "Do hand foot!") :-)
Cinnioglu et al., 2004
I made some figures for a relatively old but still important publication about the Y haplogroups observed in Turkey. For a better understanding I am showing the total and regional frequencies of the Y haplogroups.
Same numbers, just without the map:
And sorted by haplogroups:
Same numbers, just without the map:
And sorted by haplogroups:
Tuesday, April 10, 2012
Kurdish Y-DNA at 23andme Part II
Just a new update including the Yezidi.
1x E1b1b1c1a (Alevi)
1x G2a (Alevi)
2x J1 (Feyli, originally from Iran)
1x J1c3 (Sorani)
1x J2 (Kurd from Turkey)
1x J2a3a (J2a1a at 23andme; J2a4a at ISOGG 2009; he is M47+, M322+)(Yezidi)
1x T (Sorani)
1x R2a (Sorani)
1x R1b1a2* (Kurmanji from Zakho)
1x R1b1b2a (Zaza)
1x R1a1a (Sorani)
1x R1a1a (Z283+, only his paternal great-grandfather is Kurdish from Turkey)
1x I2a2a* (old I2b1*) (Sorani)
So where in the world can we find the Y haplogroup of the Yezidi individual?
Answer: Mostly in Lebanon.
Zalloua et al., 2008 (M47+):
Lebanese (140/916 = 15.3%; evenly distributed between Lebanese Druze, Muslims, and Christians)
Regueiro et al., 2006 (M47+):
Southern Iran (4/117 = 3.4%)
Abu-Amero et al., 2009 (M47+):
Saudi-Arabia (4/157 = 2.6%)
Cinnioglu et al., 2004 (M47+):
Turkey (6/523 = 1.1%)
Regional subdivision based on Cinnioglu:
a) Eastern Anatolia (Region 4): 2/82 = 2.4%
b) Central Anatolia (Region 7): 1/90 = 1.1%
c) Istanbul (Region 9): 3/81 = 3.7%
Giacomo et al., 2004 (M47+):
Syria (1/50 = 2%)
Turkey (1/168 = 0.6%)
Battaglia et al., 2009 (M47+):
Georgia (1/66 = 1.5%)
Cadenas et al., 2008 (M47+):
Qatar (1/72 = 1.4%)
UAE (3/164 = 1.8%)
Arredi et al., 2004 (M47+):
Tunisia (1/148 = 0.7%)
In the J2 Middle East Project, there are 4 confirmed cases of M47+:
M4272 Bin Eabood J2a4a
E10798 K J2a4a
169860 Saleh J2a4a
M6737 Ben Ebrahim J2a4a
In the Armenian DNA Project, there is one confirmed case of M47+:
101499 Ohanes Chantikian, Van, Armenia J2a4a J-M47
More data can be found here:
Kurdish Y-DNA Part I
Kurdish Y-DNA Part III
Kurdish Y-DNA Part IV
Kurdish Y-DNA Part V
Kurdish Y-DNA Part VI
Kurdish Y-DNA Part VII
Kurdish Y-DNA Part VIII
1x E1b1b1c1a (Alevi)
1x G2a (Alevi)
2x J1 (Feyli, originally from Iran)
1x J1c3 (Sorani)
1x J2 (Kurd from Turkey)
1x J2a3a (J2a1a at 23andme; J2a4a at ISOGG 2009; he is M47+, M322+)(Yezidi)
1x T (Sorani)
1x R2a (Sorani)
1x R1b1a2* (Kurmanji from Zakho)
1x R1b1b2a (Zaza)
1x R1a1a (Sorani)
1x R1a1a (Z283+, only his paternal great-grandfather is Kurdish from Turkey)
1x I2a2a* (old I2b1*) (Sorani)
So where in the world can we find the Y haplogroup of the Yezidi individual?
Answer: Mostly in Lebanon.
Zalloua et al., 2008 (M47+):
Lebanese (140/916 = 15.3%; evenly distributed between Lebanese Druze, Muslims, and Christians)
Regueiro et al., 2006 (M47+):
Southern Iran (4/117 = 3.4%)
Abu-Amero et al., 2009 (M47+):
Saudi-Arabia (4/157 = 2.6%)
Cinnioglu et al., 2004 (M47+):
Turkey (6/523 = 1.1%)
Regional subdivision based on Cinnioglu:
a) Eastern Anatolia (Region 4): 2/82 = 2.4%
b) Central Anatolia (Region 7): 1/90 = 1.1%
c) Istanbul (Region 9): 3/81 = 3.7%
Giacomo et al., 2004 (M47+):
Syria (1/50 = 2%)
Turkey (1/168 = 0.6%)
Battaglia et al., 2009 (M47+):
Georgia (1/66 = 1.5%)
Cadenas et al., 2008 (M47+):
Qatar (1/72 = 1.4%)
UAE (3/164 = 1.8%)
Arredi et al., 2004 (M47+):
Tunisia (1/148 = 0.7%)
In the J2 Middle East Project, there are 4 confirmed cases of M47+:
M4272 Bin Eabood J2a4a
E10798 K J2a4a
169860 Saleh J2a4a
M6737 Ben Ebrahim J2a4a
In the Armenian DNA Project, there is one confirmed case of M47+:
101499 Ohanes Chantikian, Van, Armenia J2a4a J-M47
More data can be found here:
Kurdish Y-DNA Part I
Kurdish Y-DNA Part III
Kurdish Y-DNA Part IV
Kurdish Y-DNA Part V
Kurdish Y-DNA Part VI
Kurdish Y-DNA Part VII
Kurdish Y-DNA Part VIII
mtDNA of Kurds Part III
Just a new update. For the first (as far as I know), we have some mtDNA data of an Yezidi Kurd.
He is H14.
23andme data:
1x C4b (Alevi)
1x H5a1 (Sorani)
1x H14 (Yezidi)
1x H15a (Sorani, mtDNA fully sequenced here and here)
1x H15b (Sorani)
1x HV (Sorani)
1x HV (Kurmanji from Zakho)
1x J1b (Sorani)
1x J2a1a (Kurd from Turkey)
1x N1b1 (Alevi)
1x U1a1 (Zaza)
1x U1a1 (Sorani)
1x U8b (Feyli)
So where else do we find H14?
Most known H14 cases worldwide are actually downstream of H14, i.e. H14a and H14a1, but this particular Yezidi belongs to the roots of H14. At phylotree.org, there is only one complete mtDNA sequence from Turkey (no ethnicity named) with the mtDNA haplogroup H14.
Here are the available data summarized by Ian Logan:
http://www.ianlogan.co.uk/sequences_by_group/h14_genbank_sequences.htm
H14 7645 10217
1. AM263182(Turk) Roostalu H14
H14a 16256 16352
2. AM263183(Armenia) Roostalu H14a
3. AY738951(Italy) Achilli H14a
4. EF660990(Chile) Gasparre (INCOMP) H14a
5. GQ983090(Italy) Santoro H14a
6. JN609592(Turkey) FTDNA H14a
H14a1 146 7864 11002 12870
7. EF556163 Behar2008 H14a1
Rootstalu et al., 2007:
Here is a nice map from genebase.com that summarizes occurrence of the different subclades of mtDNA haplogroup H.
He is H14.
23andme data:
1x C4b (Alevi)
1x H5a1 (Sorani)
1x H14 (Yezidi)
1x H15a (Sorani, mtDNA fully sequenced here and here)
1x H15b (Sorani)
1x HV (Sorani)
1x HV (Kurmanji from Zakho)
1x J1b (Sorani)
1x J2a1a (Kurd from Turkey)
1x N1b1 (Alevi)
1x U1a1 (Zaza)
1x U1a1 (Sorani)
1x U8b (Feyli)
So where else do we find H14?
Most known H14 cases worldwide are actually downstream of H14, i.e. H14a and H14a1, but this particular Yezidi belongs to the roots of H14. At phylotree.org, there is only one complete mtDNA sequence from Turkey (no ethnicity named) with the mtDNA haplogroup H14.
Here are the available data summarized by Ian Logan:
http://www.ianlogan.co.uk/sequences_by_group/h14_genbank_sequences.htm
H14 7645 10217
1. AM263182(Turk) Roostalu H14
H14a 16256 16352
2. AM263183(Armenia) Roostalu H14a
3. AY738951(Italy) Achilli H14a
4. EF660990(Chile) Gasparre (INCOMP) H14a
5. GQ983090(Italy) Santoro H14a
6. JN609592(Turkey) FTDNA H14a
H14a1 146 7864 11002 12870
7. EF556163 Behar2008 H14a1
Another case of H14a (not H14) is described in the Armenian DNA project:
| 190624 | H14a | 16256T, 16295T, 16352C | 150T, 263G, 309.1C, 309.2C, 315.1C |
I know of one Syriac Orthodox Assyrian with H14a as well.
Family Tree DNA has a project dedicated for H14 only.
Unfortunately, the few cases do not have any information about ethnicity,
only the 'map function' gives you a hint.Other scientific sources for mtDNA H14: In Brandstätter et al. (2006), H14 (H14a not tested) was also found Tyrol/Austria at a very low frequency (7/859 = 0.8%).
Rootstalu et al., 2007:
Monday, April 9, 2012
Y chromosome haplogroups of Kurdish people Part 2:
Today, I want to present Omer Gokcumen's publication. He analyzed the Y-STR values of 4 Central Anatolian villages that are in close proximity to each other, however, they are dominated by different ethnic groups. For some reasons (that I don't want to discuss) he used pseudonyms for the 4 villages, so please do not try to find the locations on a map. One of 4 villages is described as Kurdish, Gokcumen called this villages 'Dogukoy' in his publication.
Based on the figure 6 of the publication (see below), the following Y-STR haplogroups were observed in the Kurdish village of 'Dokukoy': E3b, I1b1, I1b2, J1, J2a, R1a, and R1b in a total of 31 tested individuals from 'Dokukoy'. E1b and J2a are dominating in the Kurdish village, while R1b, R1a, I1b1, I1b2, and J1 are present but relatively low.
A closer look at the Y-STR values of 31 tested individuals from 'Dokukoy' (can be found in the supplementary information) reveals that a lot of cases of J2a are identical or have a very narrow star-like occurence, so maybe Gokcumen et al. just tested a lot of relatives. A even more extreme pattern can be seen in the data set of the village 'Gocmenkoy' (Residents of 'Gocmenkoy' identify themselves with the Afsar clan of the Oguz tribe). Thus, I will not use any of the calculated Y-haplogroup frequencies of this publication, they are misleading. Pictures are sometimes better than 1000 words:
Green, red, blue, and yellow indicate the 'Gocmenkoy', 'Dogukoy', 'Eskikoy', and 'Merkez' settlements, respectively.
What can be used from the publication are the variation of observed Y-STR values of the Kurds of 'Dogukoy'.
I will compare these observed Y-STR haplotypes with published data and databases, maybe these 'Kurdish haplotypes' are present in other populations as well. As a starter, just the Modal haplotype for haplogroup J2a in Kurds from 'Dogukoy':
The closest matches at ysearch are (distance=6/17):
EJ7B6 (Xenofontov, Russia)
6WPPC (Y-DNA J2f-M67 MODAL HAPLOTYPE)
and
G8URM (CruzFajardo, Puerto Rico)
To be continued...
Based on the figure 6 of the publication (see below), the following Y-STR haplogroups were observed in the Kurdish village of 'Dokukoy': E3b, I1b1, I1b2, J1, J2a, R1a, and R1b in a total of 31 tested individuals from 'Dokukoy'. E1b and J2a are dominating in the Kurdish village, while R1b, R1a, I1b1, I1b2, and J1 are present but relatively low.
A closer look at the Y-STR values of 31 tested individuals from 'Dokukoy' (can be found in the supplementary information) reveals that a lot of cases of J2a are identical or have a very narrow star-like occurence, so maybe Gokcumen et al. just tested a lot of relatives. A even more extreme pattern can be seen in the data set of the village 'Gocmenkoy' (Residents of 'Gocmenkoy' identify themselves with the Afsar clan of the Oguz tribe). Thus, I will not use any of the calculated Y-haplogroup frequencies of this publication, they are misleading. Pictures are sometimes better than 1000 words:
Green, red, blue, and yellow indicate the 'Gocmenkoy', 'Dogukoy', 'Eskikoy', and 'Merkez' settlements, respectively.
What can be used from the publication are the variation of observed Y-STR values of the Kurds of 'Dogukoy'.
I will compare these observed Y-STR haplotypes with published data and databases, maybe these 'Kurdish haplotypes' are present in other populations as well. As a starter, just the Modal haplotype for haplogroup J2a in Kurds from 'Dogukoy':
| DYS 393 | DYS 390 | DYS 19 | DYS 391 | DYS 385a | DYS 385b | DYS 439 | DYS 389I | DYS 392 | DYS 389II | DYS 458 | DYS 437 | DYS 448 | DYS 456 | DYS 438 | H4 GATA | DYS 635 |
| 12 | 23 | 14 | 11 | 13 | 16 | 12 | 12 | 11 | 29 | 16 | 15 | 19 | 15 | 9 | 21 | 22 |
The closest matches at ysearch are (distance=6/17):
EJ7B6 (Xenofontov, Russia)
6WPPC (Y-DNA J2f-M67 MODAL HAPLOTYPE)
and
G8URM (CruzFajardo, Puerto Rico)
To be continued...
Friday, April 6, 2012
mtDNA of Kurds Part II
Quintana-Murci et al., 2004, analyzed the mtDNA of 20 Kurds from Iran.
I (1/20=5.0%)
W (2/20=10.0%)
HV* (2/20=10.0%)
HV1 (1/20=5.0%)
HV2 (1/20=5.0%)
H (2/20=10.0%)
U5 (1/20=5.0%)
U7 (4/20=20.0%)
U8b (2/20=10.0%)
K (1/20=5.0%)
J1 (1/20=5.0%)
I (1/20=5.0%)
W (2/20=10.0%)
HV* (2/20=10.0%)
HV1 (1/20=5.0%)
HV2 (1/20=5.0%)
H (2/20=10.0%)
U5 (1/20=5.0%)
U7 (4/20=20.0%)
U8b (2/20=10.0%)
K (1/20=5.0%)
J1 (1/20=5.0%)
Thursday, April 5, 2012
mtDNA of Kurds Part I
Today, I want to present the maternal haplogroups of Kurds. Unfortunately, information regarding the mtDNA of Kurds is very restricted.
23andme data:
1x C4b (Alevi)
1x H5a1 (Sorani)
1x H15a (Sorani, mtDNA fully sequenced here and here)
1x H15b (Sorani)
1x HV (Sorani)
1x HV (Kurmanji from Zakho)
1x J1b (Sorani)
1x J2a1a (Kurd from Turkey)
1x N1b1 (Alevi)
1x U1a1 (Zaza)
1x U1a1 (Sorani)
1x U8b (Feyli)
23andme data:
1x C4b (Alevi)
1x H5a1 (Sorani)
1x H15a (Sorani, mtDNA fully sequenced here and here)
1x H15b (Sorani)
1x HV (Sorani)
1x HV (Kurmanji from Zakho)
1x J1b (Sorani)
1x J2a1a (Kurd from Turkey)
1x N1b1 (Alevi)
1x U1a1 (Zaza)
1x U1a1 (Sorani)
1x U8b (Feyli)
Where does I2a2a-M223 (old I2b1) come from?
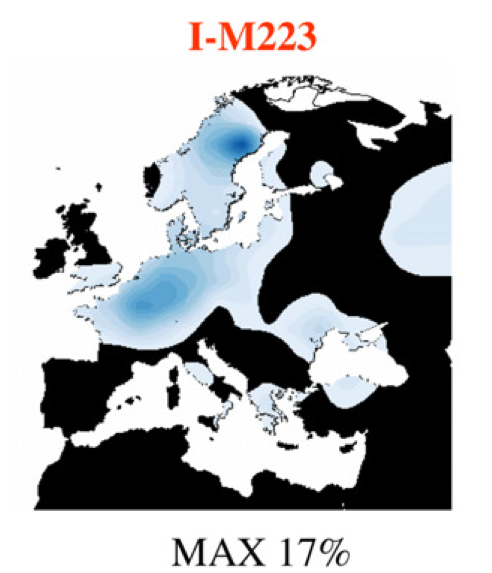
I was trying to get some information about I2b1 (M223) to understand its origin and migration. To get a first impression about the distribution of M223, below is a map based on SMGF. The peak area of I2b1 seems to be in Central Germany.
Chiaronia et al., 2009 presented a map of M223, too.
Balanovsky et al. 2008 tested for the SNP M223 and found it in several individuals from Russia.
Vologda (2/121=1.7%)
Unzha (2/52=3.8%)
Kashin (3/73=4.1%)
Livni (1/110=0.9%)
Belgorod (1/143=0.7%)
Kuban Cossacs (2/90=2.2%)
I am still collecting I2b1 data, I will update this post later today.
Edit:
In older publications M223 is also called I1c.
M223 frequency in Europe (From Table 1 in Rootsi et al., 2004):
Portuguese (5/303=1.6%)
Madeiran (Portugal) (2/132=1.5%)
Azorean (Portugal) (2/121=1.6%)
Cape Verde Islanders (1/201=0.5%)
Italian (northern Italy) (2/194=1.0%)
Italian (central Italy) (6/196=3.0%)
Italian (Calabria) (2/148=1.4%)
Italian (Albanese origin) (3/78=3.8%)
Italian (Sardinia) (2/142=1.4%)
French (Southern France) (1/38=5.3%)
French (Low Normandy) (1/42=4.8%)
French (Lyon, Poitier) (1/99=1.0%)
German (2/16=12.5%)
Dutch (3/30=10%)
Swedish (South Sweden) (6/168=3.6)
Norwegian (1/72=1.4%)
Estonian (1/210=0.5%)
Latvian (1/86=1.2%)
Polish (2/191=1.0%)
Czech and Slovak (2/198=1.0%)
Hungarian (2/162=1.2%)
Byelorussian (1/147=0.7%)
Ukrainian (3/585=0.5%)
Russian (Kostroma region) (2/53=3.7%)
Russian (Belgorod region) (1/144=0.7%)
Russian (Cossacks) (2/97=2.0%)
Russian (Adygea) (1/78=1.3%)
Mordvin (4/83=4.8%)
Chuvashes (2/80=2.5%)
Slovenian (1/55=1.8%)
Croat (mainland) (1/189=0.5%)
Albanian (4/106=3.8%)
Romanian (7/361=1.9%)
Moldavian (2/60=3.3%)
Gagauz (Moldova) (4/79=5.1%)
Greek (4/261=1.5%)
Turkey (5/741=0.7%)
Lebanese (1/66=1.5%)
The map based on Rootsi et al., 2004:
The number of tested Germans and Dutch is actually very low in this paper (N=16 and N=30, respectively), so the frequency in this region might be off.
The network of haplogroup I was obtained by using the biallelic markers and six STR loci (DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393) in 533 Hg I chromosomes from 34 populations:
Interestingly, the Near Eastern STR haplotypes of I1c (M223) and I1a (M253) are both in the 'core' of their haplogroups, respectively.
It is also interesting to note that M223 is the one of the oldest SNPs in haplogroup I.
Chiaronia et al., 2009 presented a map of M223, too.
Balanovsky et al. 2008 tested for the SNP M223 and found it in several individuals from Russia.
Vologda (2/121=1.7%)
Unzha (2/52=3.8%)
Kashin (3/73=4.1%)
Livni (1/110=0.9%)
Belgorod (1/143=0.7%)
Kuban Cossacs (2/90=2.2%)
I am still collecting I2b1 data, I will update this post later today.
Edit:
In older publications M223 is also called I1c.
M223 frequency in Europe (From Table 1 in Rootsi et al., 2004):
Portuguese (5/303=1.6%)
Madeiran (Portugal) (2/132=1.5%)
Azorean (Portugal) (2/121=1.6%)
Cape Verde Islanders (1/201=0.5%)
Italian (northern Italy) (2/194=1.0%)
Italian (central Italy) (6/196=3.0%)
Italian (Calabria) (2/148=1.4%)
Italian (Albanese origin) (3/78=3.8%)
Italian (Sardinia) (2/142=1.4%)
French (Southern France) (1/38=5.3%)
French (Low Normandy) (1/42=4.8%)
French (Lyon, Poitier) (1/99=1.0%)
German (2/16=12.5%)
Dutch (3/30=10%)
Swedish (South Sweden) (6/168=3.6)
Norwegian (1/72=1.4%)
Estonian (1/210=0.5%)
Latvian (1/86=1.2%)
Polish (2/191=1.0%)
Czech and Slovak (2/198=1.0%)
Hungarian (2/162=1.2%)
Byelorussian (1/147=0.7%)
Ukrainian (3/585=0.5%)
Russian (Kostroma region) (2/53=3.7%)
Russian (Belgorod region) (1/144=0.7%)
Russian (Cossacks) (2/97=2.0%)
Russian (Adygea) (1/78=1.3%)
Mordvin (4/83=4.8%)
Chuvashes (2/80=2.5%)
Slovenian (1/55=1.8%)
Croat (mainland) (1/189=0.5%)
Albanian (4/106=3.8%)
Romanian (7/361=1.9%)
Moldavian (2/60=3.3%)
Gagauz (Moldova) (4/79=5.1%)
Greek (4/261=1.5%)
Turkey (5/741=0.7%)
Lebanese (1/66=1.5%)
The map based on Rootsi et al., 2004:
The number of tested Germans and Dutch is actually very low in this paper (N=16 and N=30, respectively), so the frequency in this region might be off.
The network of haplogroup I was obtained by using the biallelic markers and six STR loci (DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393) in 533 Hg I chromosomes from 34 populations:
Interestingly, the Near Eastern STR haplotypes of I1c (M223) and I1a (M253) are both in the 'core' of their haplogroups, respectively.
It is also interesting to note that M223 is the one of the oldest SNPs in haplogroup I.
Monday, April 2, 2012
Kurdish Y-DNA at 23andme Part I
Today, I want to present Y chromosome results of Kurds that got analyzed by 23andme.
This is what we have so far:
1x E1b1b1c1a (Alevi)
1x G2a (Alevi)
2x J1 (Feyli, originally from Iran)
1x J1c3 (Sorani)
1x J2 (Kurd from Turkey)
1x T (Sorani)
1x R2a (Sorani)
1x R1b1a2* (Kurmanji from Zakho)
1x R1b1b2a (Zaza)
1x R1a1a (Sorani)
1x R1a1a (Z283+, only his paternal great-grandfather is Kurdish from Turkey)
1x I2a2a (old I2b1) (Sorani)
Luckily, the last three in this list went a little bit further and got a more detailed analysis of their Y chromosome at FTDNA.
R1a1a (Sorani):
The Sorani with R1a1a turns out to be Z93+ L342.2+ L657-, a branch of R1a1a that is currently called R1a1a1h1a, but the nomenclature can change. He is H1483 and can be found in the 'R1a1a and Subclades Y chromosome project', a project that is designated 'to understand the haplogroup and explore the deeper structure of R1a1a'.
Individuals with Z93+ L342.2+ L657- are mostly from the Middle East, Central Asia, and South Asia. Additionally, Ashkenazi Jews with R1a1a are mostly positive for the two SNPs Z93 and L342.2 and negative for L657. In the 'R1a1a and Subclades Y chromosome project', this subclade is called 2C.
R1a1a (Z283+, only his paternal great-grandfather is Kurdish from Turkey):
The second individual with paternal Kurdish ancestry and the Y chromosome haplogroup R1a1a is 214352 in the same project. He is definitely a very interesting case for Kurds as well as for the 'R1a1a and Subclades Y chromosome project' itself, since he belongs to a subclade (called subclade 3 in the project=R1a1a1g*) that was just recently discovered and that split from the most common European subclades in very ancient times. He is positive for Z283 and negative for all known underlying SNPs: Z283+ M417+ Z93- Z280- Pk5- P98- M64.2- M56- M458- M434- M334- M157.1- L260- L176.1- L175-.
To get a better idea of R1a1a and its subclades (lineage tree image from the project):
I2a2a-M223 (old name I2b1) (Sorani):
The third individual that provided more detailed information about his Y chromosome haplogroup is 229546. He can be found in the I2b1/M223 Y-CLAN STUDY, a project that is focusing on individuals that are positive for the SNP M223. In general, M223+ people were mostly found in Germany and Germanic speaking countries, however, there are a few cases described elsewhere.
The interesting part about the Kurdish Sorani 229546 is that he does not belong to one of the subclades of I2b1 (that emerged later in time) but he belongs to the roots of this haplogroup. His closest match so far is an Armenian individual (Distance 14/67; Shahnazar alias USC2T at ysearch).
R1b1a2* (R1b1b2 based on 23andme; Kurmanji from Zakho):
Although this individual did not get a more detailed analysis of his Y chromosome, we can say the following:
His haplogroup is old, older than the European subclades of R1b. His subclade is very rare and it is the root for the European R1b.
Note: The nomenclature is sometimes confusing, the most reliable source is ISOGG. 23andme tends to use the older nomenclature (in this case R1b1b2).
More data can be found here:
Kurdish Y-DNA Part II
Kurdish Y-DNA Part III
Kurdish Y-DNA Part IV
Kurdish Y-DNA Part V
Kurdish Y-DNA Part VI
Kurdish Y-DNA Part VII
Kurdish Y-DNA Part VIII
This is what we have so far:
1x E1b1b1c1a (Alevi)
1x G2a (Alevi)
2x J1 (Feyli, originally from Iran)
1x J1c3 (Sorani)
1x J2 (Kurd from Turkey)
1x T (Sorani)
1x R2a (Sorani)
1x R1b1a2* (Kurmanji from Zakho)
1x R1b1b2a (Zaza)
1x R1a1a (Sorani)
1x R1a1a (Z283+, only his paternal great-grandfather is Kurdish from Turkey)
1x I2a2a (old I2b1) (Sorani)
Luckily, the last three in this list went a little bit further and got a more detailed analysis of their Y chromosome at FTDNA.
R1a1a (Sorani):
The Sorani with R1a1a turns out to be Z93+ L342.2+ L657-, a branch of R1a1a that is currently called R1a1a1h1a, but the nomenclature can change. He is H1483 and can be found in the 'R1a1a and Subclades Y chromosome project', a project that is designated 'to understand the haplogroup and explore the deeper structure of R1a1a'.
Individuals with Z93+ L342.2+ L657- are mostly from the Middle East, Central Asia, and South Asia. Additionally, Ashkenazi Jews with R1a1a are mostly positive for the two SNPs Z93 and L342.2 and negative for L657. In the 'R1a1a and Subclades Y chromosome project', this subclade is called 2C.
R1a1a (Z283+, only his paternal great-grandfather is Kurdish from Turkey):
The second individual with paternal Kurdish ancestry and the Y chromosome haplogroup R1a1a is 214352 in the same project. He is definitely a very interesting case for Kurds as well as for the 'R1a1a and Subclades Y chromosome project' itself, since he belongs to a subclade (called subclade 3 in the project=R1a1a1g*) that was just recently discovered and that split from the most common European subclades in very ancient times. He is positive for Z283 and negative for all known underlying SNPs: Z283+ M417+ Z93- Z280- Pk5- P98- M64.2- M56- M458- M434- M334- M157.1- L260- L176.1- L175-.
To get a better idea of R1a1a and its subclades (lineage tree image from the project):
I2a2a-M223 (old name I2b1) (Sorani):
The third individual that provided more detailed information about his Y chromosome haplogroup is 229546. He can be found in the I2b1/M223 Y-CLAN STUDY, a project that is focusing on individuals that are positive for the SNP M223. In general, M223+ people were mostly found in Germany and Germanic speaking countries, however, there are a few cases described elsewhere.
The interesting part about the Kurdish Sorani 229546 is that he does not belong to one of the subclades of I2b1 (that emerged later in time) but he belongs to the roots of this haplogroup. His closest match so far is an Armenian individual (Distance 14/67; Shahnazar alias USC2T at ysearch).
R1b1a2* (R1b1b2 based on 23andme; Kurmanji from Zakho):
Although this individual did not get a more detailed analysis of his Y chromosome, we can say the following:
His haplogroup is old, older than the European subclades of R1b. His subclade is very rare and it is the root for the European R1b.
Note: The nomenclature is sometimes confusing, the most reliable source is ISOGG. 23andme tends to use the older nomenclature (in this case R1b1b2).
More data can be found here:
Kurdish Y-DNA Part II
Kurdish Y-DNA Part III
Kurdish Y-DNA Part IV
Kurdish Y-DNA Part V
Kurdish Y-DNA Part VI
Kurdish Y-DNA Part VII
Kurdish Y-DNA Part VIII
Subscribe to:
Posts (Atom)